Publications
Highlights
(For a full list see Google Scholar).
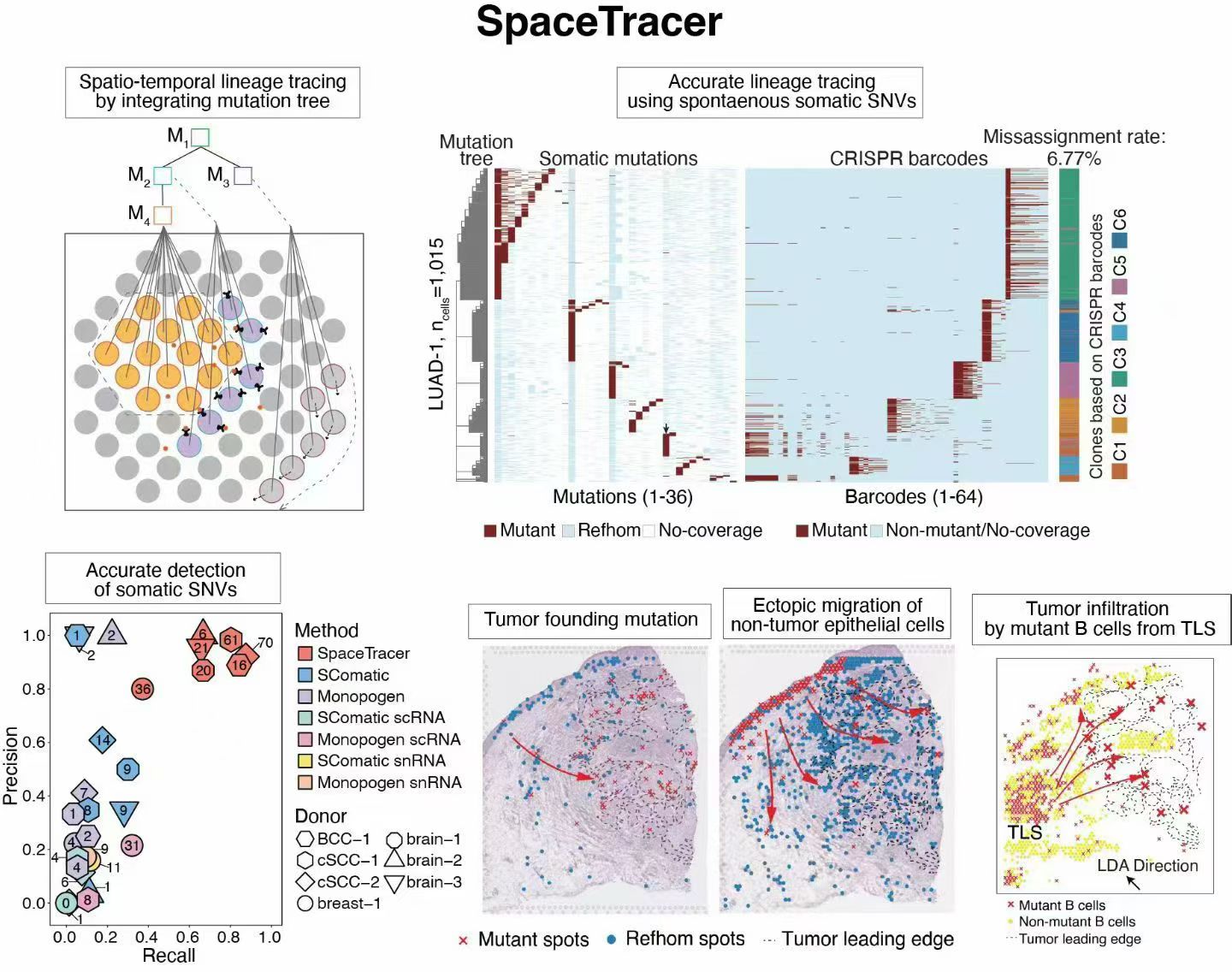
Spatial transcriptomics reveals tissue architecture but lacks lineage-tracing capability in humans. We developed SpaceTracer, a computational framework that detects somatic SNVs directly from spatial transcriptomics data and reconstructs cellular phylogenies in situ. Using this approach, we traced tumor initiation and cell migration in human cutaneous squamous cell carcinoma and uncovered lineage-associated transcriptional and spatial dynamics. SpaceTracer provides a perturbation-free strategy for high-resolution spatiotemporal lineage tracing in complex tissues.
Yang, Z. *, Yao, M. *, Yang, Q. *, Du, Y. *, Lu, J. *, Wu, X., Lin, J., Qian, Z., Hu, S., Xia, Y., Liu, H., Zhou, Q., Ma, X., Luo, Y., Fan, W., Pei, W., Xia, Y., Yu, X., Luan, J., Zhang, Q., Zhang, Y., Wang, Q., Zeng, J., Zhang, Y., Wu, W.#, Dou, Y.#
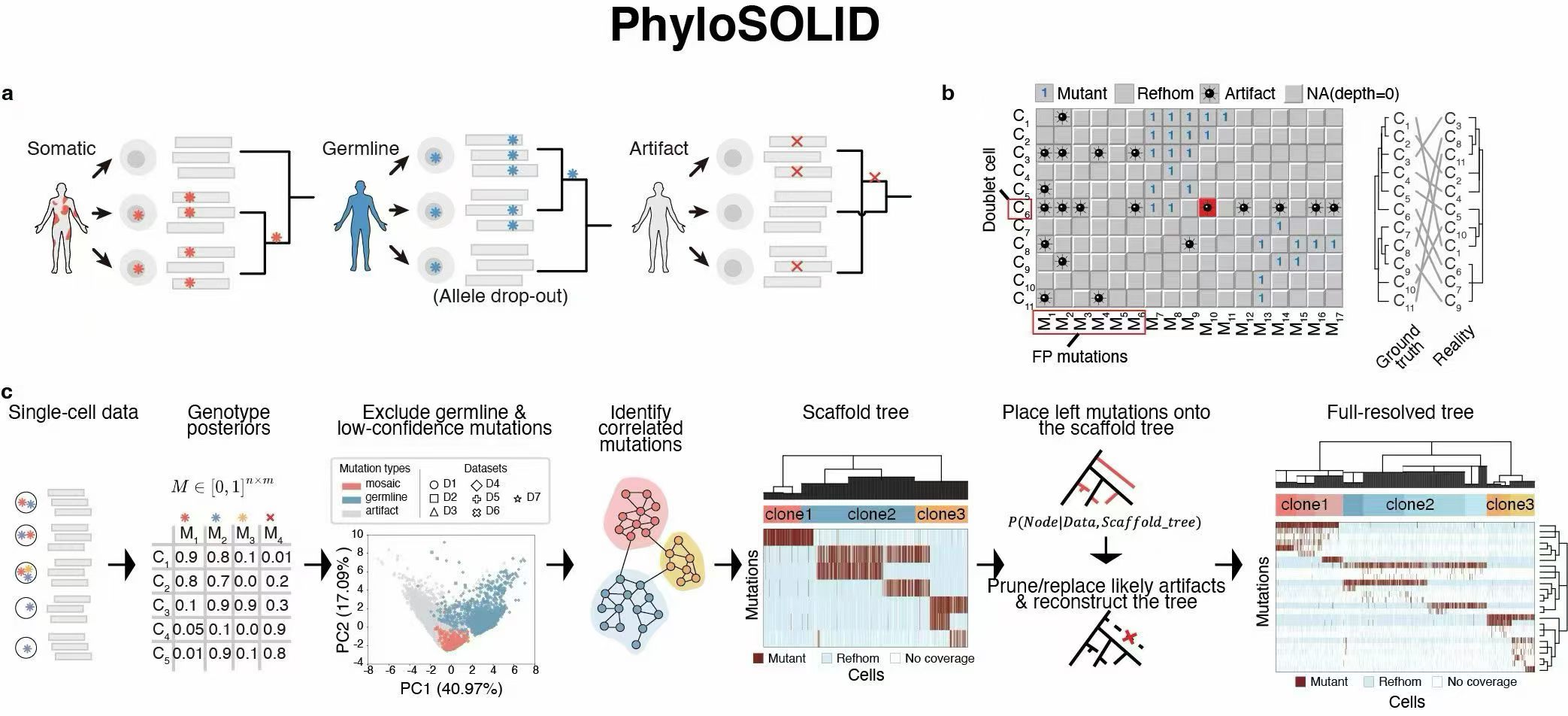
Lineage reconstruction from single-cell sequencing data is often confounded by high error rates and sparse mutation signals. We developed PhyloSOLID, a phylogenetic framework that builds a high-confidence backbone tree from reliable mutations and progressively refines it using Bayesian modeling. Benchmarking on simulated and real datasets demonstrates improved accuracy for lineage reconstruction from both single-cell RNA-seq and DNA-seq data. PhyloSOLID enables more reliable decoding of cellular evolutionary histories in development and disease.
Yang, Q. *, Liu, Y. *, Yang, J., Wu, X., Yang, Z., Xia, Y., Zheng, Y., Lu, J., Yao, M., Du, Y., Liu, H., Li, N.#, Dou, Y.#
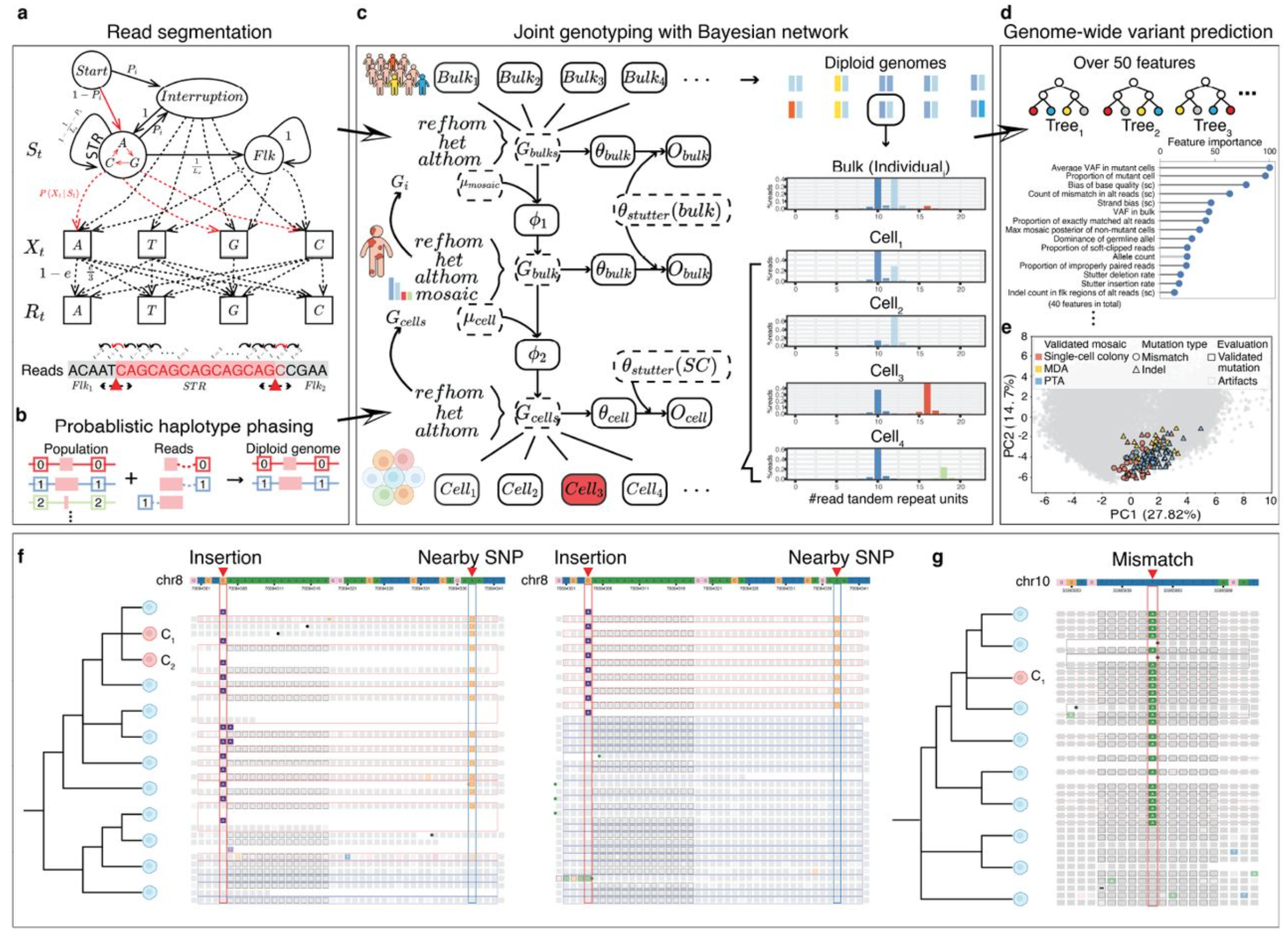
Short tandem repeats (STRs) are highly mutable genomic elements implicated in gene regulation and disease, yet their mosaic mutations are difficult to detect at single-cell resolution. We developed BayesMonSTR, a computational method for identifying mosaic STR mutations genome-wide from single-cell sequencing data. Applying this approach revealed age-associated accumulation of STR insertions and deletions, particularly in neurons of the human prefrontal cortex. These mutations are enriched at transcription start sites and active enhancers, revealing a previously underexplored landscape of mosaic STR variation.
Wang, C. *, Fan, W. *, Wang, W. *, Xia, Y. *, Lu, J. *, Ma, X. *, Yu, J. *, Zheng, Y., Luo, Y., Li, W., Yang, Q., Lin, M., Liu, H., Lan, Y., Li, C., Liu, X., He, D., Cai, S., Yu, X., Zhou, D., Kellis, M., Xiong, X., Xie, Q.#, Dou, Y.#
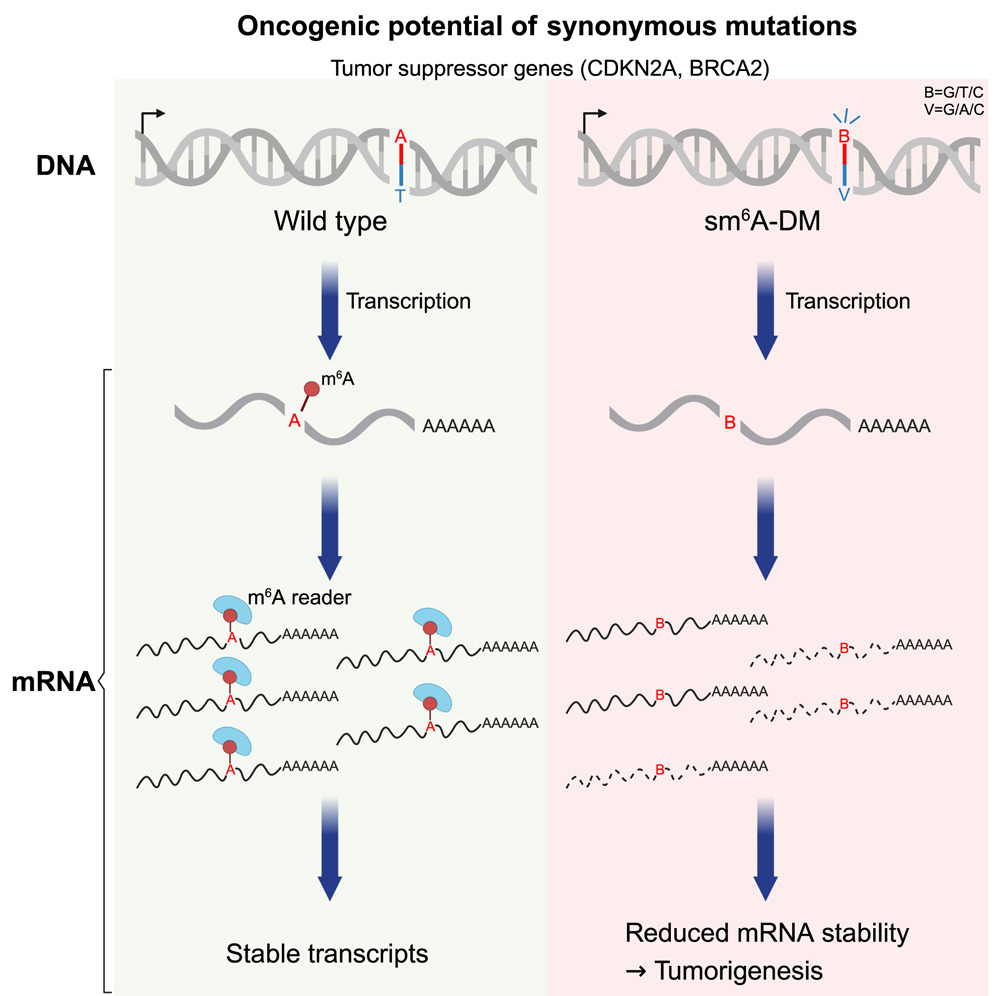
Synonymous mutations, which do not alter the amino acid sequence of proteins, were once considered functionally silent. This study reveals they promote tumorigenesis by disrupting N6-methyladenosine (m6A) modifications—the most prevalent eukaryotic mRNA modification—thereby destabilizing tumor suppressor mRNAs. Key findings include enrichment of synonymous m6A-disrupting mutations in tumor suppressor genes and their role in sensitizing tumors to targeted therapies.
Lan, Y.*, Xia, Z.*, Shao, Q.*, Lin, P., Lu, J., Xiao, X., Zheng, M., Chen, D.#, Dou, Y.#, Xie, Q.#
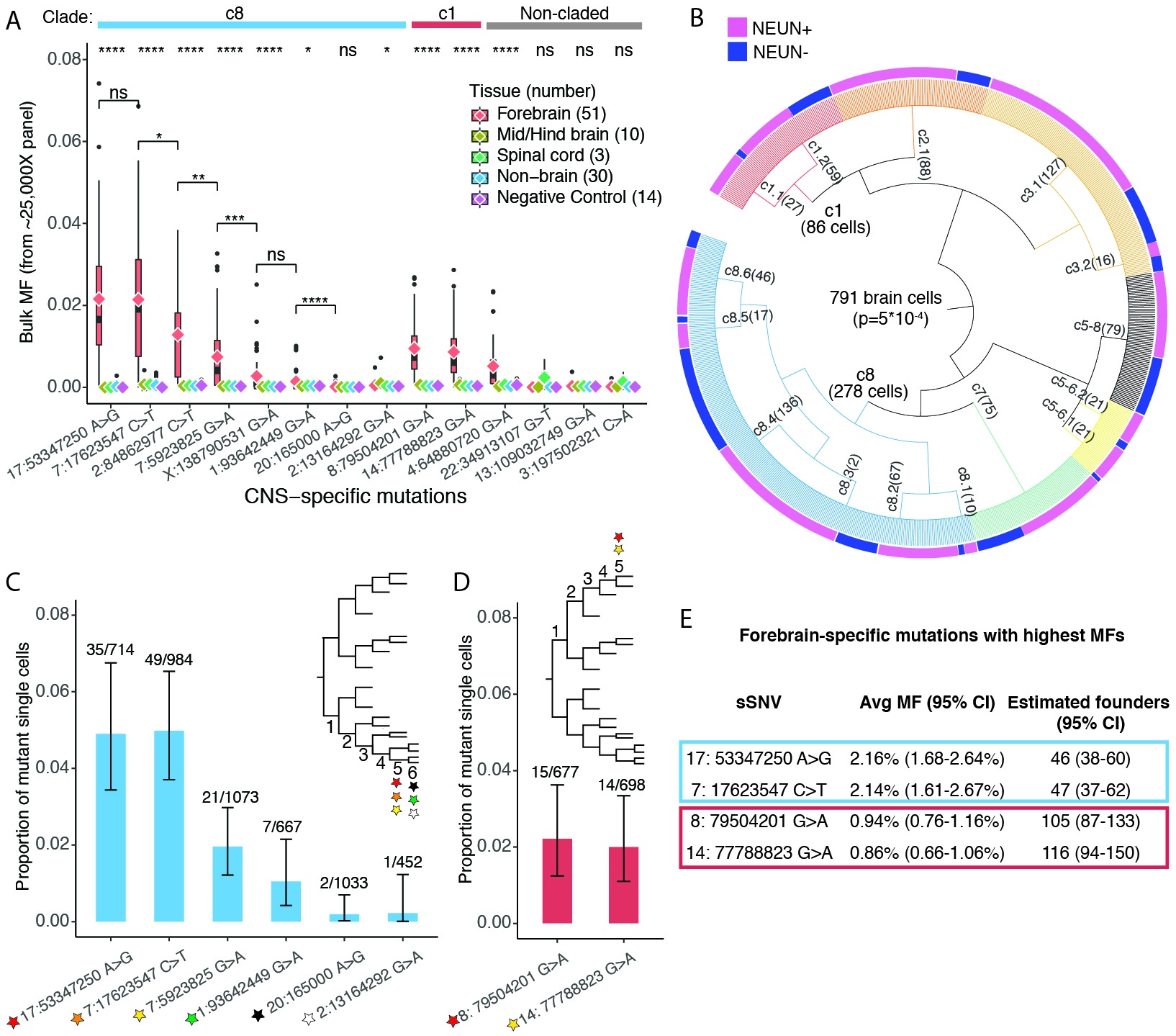
Cell lineage information is fundamental to understanding organismal development, but very little direct information is available for humans. By using mosaic mutations as endogeneous markers, we demonstrated asymmetric contributions of early progenitors to extraembryonic tissues, distinct germ layers, and organs. Our data also suggest onset of gastrulation at an effective progenitor pool of about 170 cells and about 50 to 100 founders for the forebrain.
Bizzotto, S.*, Dou, Y.*, Ganz, J.*, Doan, R. N., Kwon, M., Bohrson, C. L., Kim, S. N., Bae, T., Abyzov, A., Nimh Brain Somatic Mosaicism Network, Park, P. J.#, Walsh, C. A.#
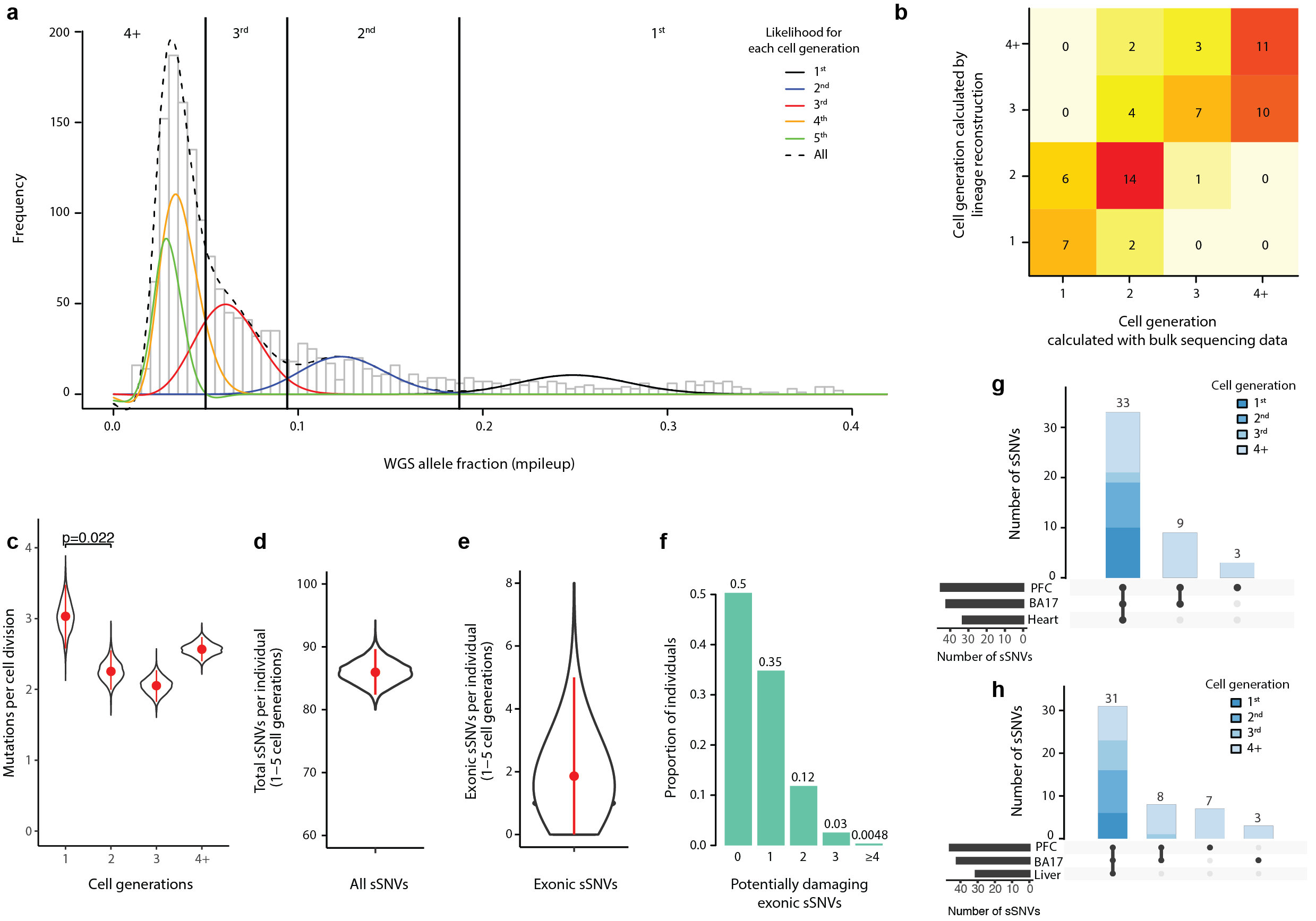
Our analysis of mosaic mutations in non-cancer individuals reveals that the first cell division after fertilization produces ~3.4 mutations, followed by 2–3 mutations in subsequent generations. This suggests that a typical individual possesses ~80 somatic single-nucleotide variants present in ≥2% of cells—comparable to the number of de novo germline mutations per generation—with about half of individuals having at least one potentially function-altering somatic mutation somewhere in the cortex.
Rodin, R. E.*, Dou, Y.*, Kwon, M., Sherman, M. A., D’Gama, A. M., Doan, R. N., Rento, L. M., Girskis, K. M., Bohrson, C. L., Kim, S. N., Nadig, A., Luquette, L. J., Gulhan, D. C., Brain Somatic Mosaicism, Network, Park, P. J.#, Walsh, C. A.#
Nat Neurosci, 1546-1726 (2021)
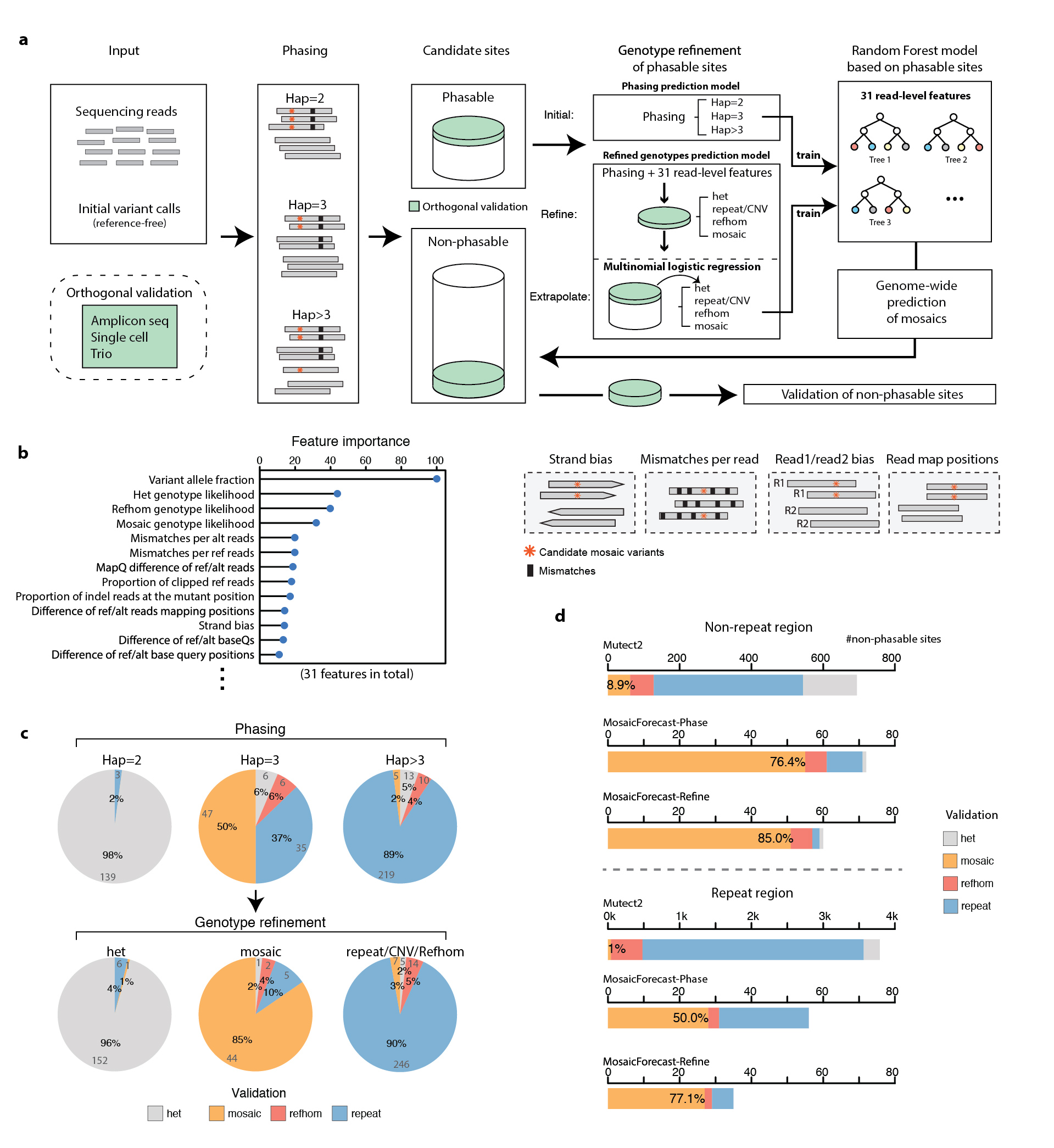
Detection of mosaic mutations that arise in normal development is challenging, as such mutations are typically present in only a minute fraction of cells and there is no clear matched control for removing germline variants and systematic artifacts. We present MosaicForecast, a machine-learning method that leverages read-based phasing and read-level features to accurately detect mosaic single-nucleotide variants and indels, achieving a multifold increase in specificity compared with existing algorithms. Using single-cell sequencing and targeted sequencing, we validated 80–90% of the mosaic single-nucleotide variants and 60–80% of indels detected in human brain whole-genome sequencing data. Our method should help elucidate the contribution of mosaic somatic mutations to the origin and development of disease.
Dou, Y., Kwon, M., Rodin, R. E., Cortes-Ciriano, I., Doan, R., Luquette, L. J., Galor, A., Bohrson, C., Walsh, C. A., Park, P. J.
Nat Biotechnol, 38(3), 314-319 (2020)
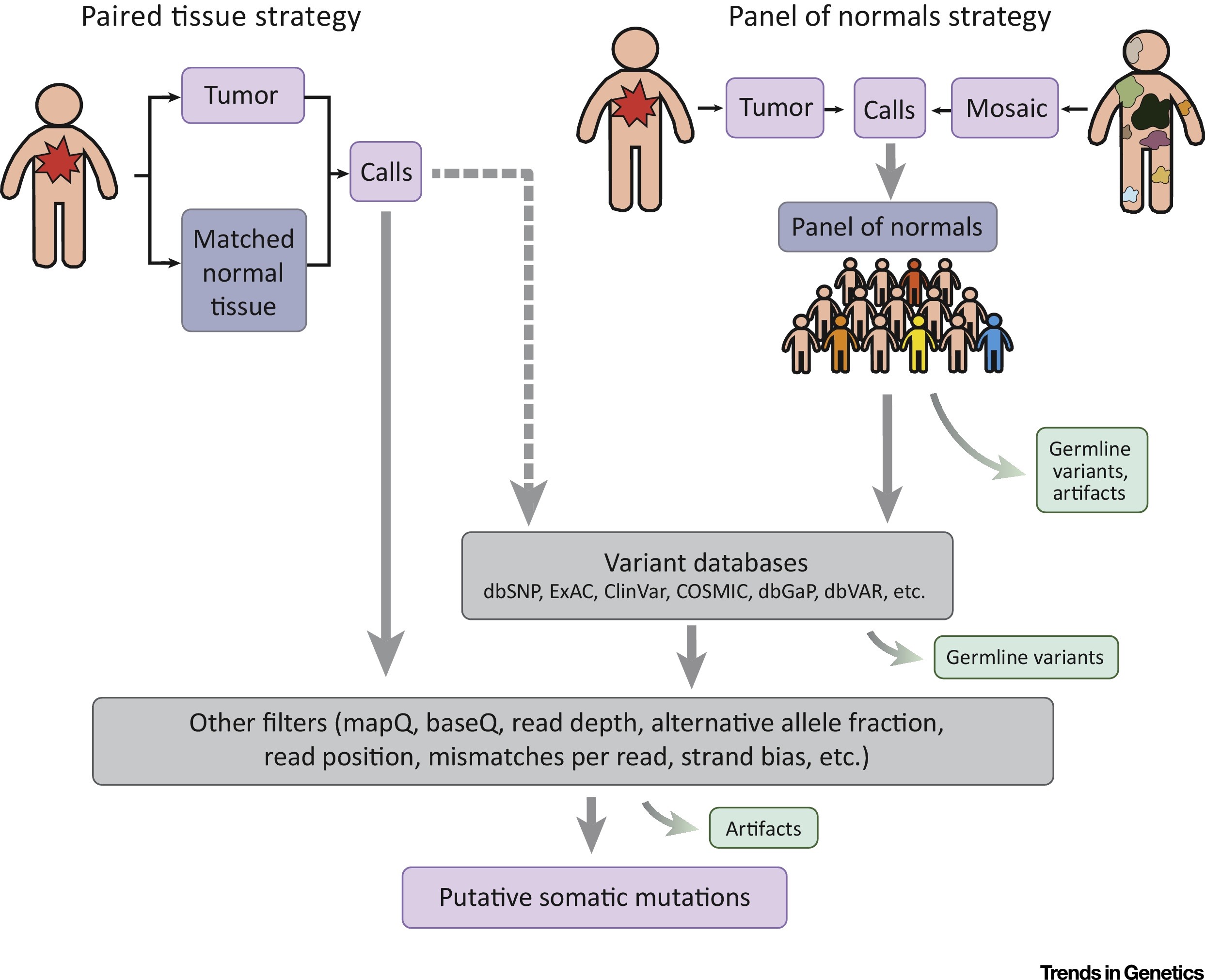
We describe here approaches for characterizing somatic mutations in normal and non-tumor disease tissues. We discuss several experimental designs and common pitfalls in somatic mutation detection, as well as more recent developments such as phasing and linked-read technology. With the dramatically increasing numbers of samples undergoing genome sequencing, bioinformatic analysis will enable the characterization of somatic mutations and their impact on non-cancer tissues.
Dou, Y.*, Gold, H. D.*, Luquette, L. J.*, Park, P. J.
Trends Genet, 34(7), 545-557 (2018)

We estimated that point mosaic mutatinos in probands or de novo mutations inherited from parental point mosaic mutations increased the risk of ASD by approximately 6%. Adding mosaic mutations into the transmission and de novo association test model revealed 13 new ASD risk genes. These results expand the existing repertoire of genes involved in ASD and shed new light on the contribution of genomic mosaicisms to ASD diagnoses and autistic traits.
Dou, Y., Yang, X., Li, Z., Wang, S., Zhang, Z., Ye, A. Y., Yan, L., Yang, C., Wu, Q., Li, J., Zhao, B., Huang, A. Y.#, Wei, L.#
Hum Mutat, 38(8), 1002-1013 (2017)